R for SQListas, what’s that about?
This is the 2-part blog version of a talk I’ve given at DOAG Conference this week. I’ve also uploaded the slides (no ppt; just pretty R presentation 😉 ) to the articles section, but if you’d like a little text I’m encouraging you to read on. That is, if you’re in the target group for this post/talk.
For this post, let me assume you’re a SQL girl (or guy). With SQL you’re comfortable (an expert, probably), you know how to get and manipulate your data, no nesting of subselects has you scared ;-). And now there’s this R language people are talking about, and it can do so many things they say, so you’d like to make use of it too – so now does this mean you have to start from scratch and learn – not only a new language, but a whole new paradigm? Turns out … ok. So that’s the context for this post.
Let’s talk about the weather
So in this post, I’d like to show you how nice R is to use if you come from SQL. But this isn’t going to be a syntax-only post. We’ll be looking at real datasets and trying to answer a real question.
Personally I’m very interested in how the weather’s going to develop in the future, especially in the nearer future, and especially regarding the area where I live (I know. It’s egocentric.). Specifically, what worries me are warm winters, and I’ll be clutching to any straw that tells me it’s not going to get warmer still 😉
So I’ve downloaded / prepared two datasets, both climate / weather-related. The first is the average global temperatures dataset from the Berkeley Earth Surface Temperature Study, nicely packaged by Kaggle (a website for data science competitions; https://www.kaggle.com/berkeleyearth/climate-change-earth-surface-t…). This contains measurements from 1743 on, up till 2013. The monthly averages have been obtained using sophisticated scientific procedures available on the Berkeley Earth website (http://berkeleyearth.org/).
The second is daily weather data for Munich, obtained from www.wunderground.com. This dataset was retrieved manually, and the period was chosen so as to not contain too many missing values. The measurements range from 1997 to 2015, and have been aggregated by taking a monthly average.
Let’s start our journey through R land, reading in and looking at the beginning of the first dataset:
library(tidyverse)
library(lubridate)
df <- read_csv('data/GlobalLandTemperaturesByCity.csv')
head(df)
df <- read_csv('data/GlobalLandTemperaturesByCity.csv')
head(df)
## 1 1743-11-01 6.068 1.737 Århus
## 2 1743-12-01 NA NA Århus
## 3 1744-01-01 NA NA Århus
## 4 1744-02-01 NA NA Århus
## 5 1744-03-01 NA NA Århus
## 6 1744-04-01 5.788 3.624 Århus
## # … with 3 more variables: Country , Latitude ,
## # Longitude
Now we’d like to explore the dataset. With SQL, this is easy: We use WHERE to filter rows, SELECT to select columns, GROUP BY to aggregate by one or more variables…And of course, we often need to JOIN tables, and sometimes, perform set operations. Then there’s all kinds of analytic functions, such as LAG() and LEAD(). How do we do all this in R?
Entering the tidyverse
Luckily for the SQLista, writing elegant, functional, and often rather SQL-like code in R is easy. All we need to do is … enter the tidyverse. Actually, we’ve already entered it – doing library(tidyverse) – and used it to read in our csv file (read_csv)!
The tidyverse is a set of packages, developed by Hadley Wickham, Chief Scientist at Rstudio, designed to make working with R easier and more consistent (and more fun). We load data from files using readr, clean up datasets that are not in third normal form using tidyr, manipulate data with dplyr, and plot them with ggplot2.
For our task of data exploration, it is dplyr we need. Before we even begin, let’s rename the columns so they have shorter names:
df <- rename(df, avg_temp = AverageTemperature, avg_temp_95p = AverageTemperatureUncertainty, city = City, country = Country, lat = Latitude, long = Longitude)
head(df)
## # A tibble: 6 × 7
## dt avg_temp avg_temp_95p city country lat long
##
## 1 1743-11-01 6.068 1.737 Århus Denmark 57.05N 10.33E
## 2 1743-12-01 NA NA Århus Denmark 57.05N 10.33E
## 3 1744-01-01 NA NA Århus Denmark 57.05N 10.33E
## 4 1744-02-01 NA NA Århus Denmark 57.05N 10.33E
## 5 1744-03-01 NA NA Århus Denmark 57.05N 10.33E
## 6 1744-04-01 5.788 3.624 Århus Denmark 57.05N 10.33E
distinct() (SELECT DISTINCT)
Good. Now that we have this new dataset containing temperature measurements, really the first thing we want to know is: What locations (countries, cities) do we have measurements for?
To find out, just do distinct():
distinct(df, country)
## # A tibble: 159 × 1
## country
##
## 1 Denmark
## 2 Turkey
## 3 Kazakhstan
## 4 China
## 5 Spain
## 6 Germany
## 7 Nigeria
## 8 Iran
## 9 Russia
## 10 Canada
## # … with 149 more rows
distinct(df, city)
## # A tibble: 3,448 × 1
## city
##
## 1 Århus
## 2 Çorlu
## 3 Çorum
## 4 Öskemen
## 5 Ürümqi
## 6 A Coruña
## 7 Aachen
## 8 Aalborg
## 9 Aba
## 10 Abadan
## # … with 3,438 more rows
filter() (WHERE)
OK. Now as I said I’m really first and foremost curious about measurements from Munich, so I’ll have to restrict the rows. In SQL I’d need a WHERE clause, in R the equivalent is filter():
filter(df, city == 'Munich')
## # A tibble: 3,239 × 7
## dt avg_temp avg_temp_95p city country lat long
##
## 1 1743-11-01 1.323 1.783 Munich Germany 47.42N 10.66E
## 2 1743-12-01 NA NA Munich Germany 47.42N 10.66E
## 3 1744-01-01 NA NA Munich Germany 47.42N 10.66E
## 4 1744-02-01 NA NA Munich Germany 47.42N 10.66E
## 5 1744-03-01 NA NA Munich Germany 47.42N 10.66E
## 6 1744-04-01 5.498 2.267 Munich Germany 47.42N 10.66E
## 7 1744-05-01 7.918 1.603 Munich Germany 47.42N 10.66E
This is how we combine conditions if we have more than one of them in a where clause:
# AND
filter(df, city == 'Munich', year(dt) > 2000)
## # A tibble: 153 × 7
## dt avg_temp avg_temp_95p city country lat long
##
## 1 2001-01-01 -3.162 0.396 Munich Germany 47.42N 10.66E
## 2 2001-02-01 -1.221 0.755 Munich Germany 47.42N 10.66E
## 3 2001-03-01 3.165 0.512 Munich Germany 47.42N 10.66E
## 4 2001-04-01 3.132 0.329 Munich Germany 47.42N 10.66E
## 5 2001-05-01 11.961 0.150 Munich Germany 47.42N 10.66E
## 6 2001-06-01 11.468 0.377 Munich Germany 47.42N 10.66E
## 7 2001-07-01 15.037 0.316 Munich Germany 47.42N 10.66E
## 8 2001-08-01 15.761 0.325 Munich Germany 47.42N 10.66E
## 9 2001-09-01 7.897 0.420 Munich Germany 47.42N 10.66E
## 10 2001-10-01 9.361 0.252 Munich Germany 47.42N 10.66E
## # ... with 143 more rows
# OR
filter(df, city == ‘Munich’ | year(dt) > 2000)
## # A tibble: 540,116 × 7
## dt avg_temp avg_temp_95p city country lat long
##
## 1 2001-01-01 1.918 0.381 Århus Denmark 57.05N 10.33E
## 2 2001-02-01 0.241 0.328 Århus Denmark 57.05N 10.33E
## 3 2001-03-01 1.310 0.236 Århus Denmark 57.05N 10.33E
## 4 2001-04-01 5.890 0.158 Århus Denmark 57.05N 10.33E
## 5 2001-05-01 12.016 0.351 Århus Denmark 57.05N 10.33E
## 6 2001-06-01 13.944 0.352 Århus Denmark 57.05N 10.33E
## 7 2001-07-01 18.453 0.367 Århus Denmark 57.05N 10.33E
## 8 2001-08-01 17.396 0.287 Århus Denmark 57.05N 10.33E
## 9 2001-09-01 13.206 0.207 Århus Denmark 57.05N 10.33E
## 10 2001-10-01 11.732 0.200 Århus Denmark 57.05N 10.33E
## # … with 540,106 more rows
select() (SELECT)
Now, often we don’t want to see all the columns/variables. In SQL we SELECT what we’re interested in, and it’s select() in R, too:
select(filter(df, city == 'Munich'), avg_temp, avg_temp_95p)
## # A tibble: 3,239 × 2
## avg_temp avg_temp_95p
##
## 1 1.323 1.783
## 2 NA NA
## 3 NA NA
## 4 NA NA
## 5 NA NA
## 6 5.498 2.267
## 7 7.918 1.603
## 8 11.070 1.584
## 9 12.935 1.653
## 10 NA NA
## # … with 3,229 more rows
arrange() (ORDER BY)
How about ordered output? This can be done using arrange():
arrange(select(filter(df, city == 'Munich'), dt, avg_temp), avg_temp)
## # A tibble: 3,239 × 2
## dt avg_temp
##
## 1 1956-02-01 -12.008
## 2 1830-01-01 -11.510
## 3 1767-01-01 -11.384
## 4 1929-02-01 -11.168
## 5 1795-01-01 -11.019
## 6 1942-01-01 -10.785
## 7 1940-01-01 -10.643
## 8 1895-02-01 -10.551
## 9 1755-01-01 -10.458
## 10 1893-01-01 -10.381
## # … with 3,229 more rows
Do you think this is starting to get difficult to read? What if we add FILTER and GROUP BY operations to this query? Fortunately, with dplyr it is possible to avoid paren hell as well as stepwise assignment using the pipe operator, %>%.
Meet: %>% – the pipe
The pipe transforms an expression of form x %>% f(y) into f(x, y) and so, allows us write the above operation like this:
df %>% filter(city == 'Munich') %>% select(dt, avg_temp) %>% arrange(avg_temp)
This looks a lot like the fluent API design popular in some object oriented languages, or the bind operator, >>=, in Haskell.
It also looks a lot more like SQL. However, keep in mind that while SQL is declarative, the order of operations matters when you use the pipe (as the name says, the output of one operation is piped to another). You cannot, for example, write this (trying to emulate SQL‘s SELECT – WHERE – ORDER BY ): df %>% select(dt, avg_temp) %>% filter(city == ‘Munich’) %>% arrange(avg_temp). This can’t work because after a new dataframe has been returned from the select, the column city is not longer available.
arrange() (GROUP BY)
Now that we’ve introduced the pipe, on to group by. This is achieved in dplyr using group_by() (for grouping, obviously) and summarise() for aggregation.
Let’s find the countries we have most – and least, respectively – records for:
# most records
df %>% group_by(country) %>% summarise(count=n()) %>% arrange(count %>% desc())
## # A tibble: 159 × 2
## country count
##
## 1 India 1014906
## 2 China 827802
## 3 United States 687289
## 4 Brazil 475580
## 5 Russia 461234
## 6 Japan 358669
## 7 Indonesia 323255
## 8 Germany 262359
## 9 United Kingdom 220252
## 10 Mexico 209560
## # … with 149 more rows
# least records
df %>% group_by(country) %>% summarise(count=n()) %>% arrange(count)
## # A tibble: 159 × 2
## country count
##
## 1 Papua New Guinea 1581
## 2 Oman 1653
## 3 Djibouti 1797
## 4 Eritrea 1797
## 5 Botswana 1881
## 6 Lesotho 1881
## 7 Namibia 1881
## 8 Swaziland 1881
## 9 Central African Republic 1893
## 10 Congo 1893
How about finding the average, minimum and maximum temperatures per month, looking at just records from Germany, and that originate after 1949?
df %>% filter(country == 'Germany', !is.na(avg_temp), year(dt) > 1949) %>% group_by(month(dt)) %>% summarise(count = n(), avg = mean(avg_temp), min = min(avg_temp), max = max(avg_temp))
## # A tibble: 12 × 5
## `month(dt)` count avg min max
##
## 1 1 5184 0.3329331 -10.256 6.070
## 2 2 5184 1.1155843 -12.008 7.233
## 3 3 5184 4.5513194 -3.846 8.718
## 4 4 5184 8.2728137 1.122 13.754
## 5 5 5184 12.9169965 5.601 16.602
## 6 6 5184 15.9862500 9.824 21.631
## 7 7 5184 17.8328285 11.697 23.795
## 8 8 5184 17.4978752 11.390 23.111
## 9 9 5103 14.0571383 7.233 18.444
## 10 10 5103 9.4110645 0.759 13.857
## 11 11 5103 4.6673114 -2.601 9.127
## 12 12 5103 1.3649677 -8.483 6.217
In this way, aggregation queries can be written that are powerful and very readable at the same time. So at this point, we know how to do basic selects with filtering and grouping. How about joins?
JOINs
Dplyr provides inner_join(), left_join(), right_join() and full_join() operations, as well as semi_join() and anti_join(). From the SQL viewpoint, these work exactly as expected.
To demonstrate a join, we’ll now load the second dataset, containing daily weather data for Munich, and aggregate it by month:
daily_1997_2015 % summarise(mean_temp = mean(mean_temp))
monthly_1997_2015
## # A tibble: 228 × 2
## month mean_temp
##
## 1 1997-01-01 -3.580645
## 2 1997-02-01 3.392857
## 3 1997-03-01 6.064516
## 4 1997-04-01 6.033333
## 5 1997-05-01 13.064516
## 6 1997-06-01 15.766667
## 7 1997-07-01 16.935484
## 8 1997-08-01 18.290323
## 9 1997-09-01 13.533333
## 10 1997-10-01 7.516129
## # … with 218 more rows
Fine. Now let’s join the two datasets on the date column (their respective keys), telling R that this column is named dt in one dataframe, month in the other:
df % select(dt, avg_temp) %>% filter(year(dt) > 1949)
df %>% inner_join(monthly_1997_2015, by = c("dt" = "month"), suffix )
## # A tibble: 705,510 × 3
## dt avg_temp mean_temp
##
## 1 1997-01-01 -0.742 -3.580645
## 2 1997-02-01 2.771 3.392857
## 3 1997-03-01 4.089 6.064516
## 4 1997-04-01 5.984 6.033333
## 5 1997-05-01 10.408 13.064516
## 6 1997-06-01 16.208 15.766667
## 7 1997-07-01 18.919 16.935484
## 8 1997-08-01 20.883 18.290323 of perceptrons
## 9 1997-09-01 13.920 13.533333
## 10 1997-10-01 7.711 7.516129
## # … with 705,500 more rows
As we see, average temperatures obtained for the same month differ a lot from each other. Evidently, the methods of averaging used (by us and by Berkeley Earth) were very different. We will have to use every dataset separately for exploration and inference.
Set operations
Having looked at joins, on to set operations. The set operations known from SQL can be performed using dplyr’s intersect(), union(), and setdiff() methods. For example, let’s combine the Munich weather data from before 2016 and from 2016 in one data frame:
daily_2016 % arrange(day)
## # A tibble: 7,195 × 23
## day max_temp mean_temp min_temp dew mean_dew min_dew max_hum
##
## 1 1997-01-01 -8 -12 -16 -13 -14 -17 92
## 2 1997-01-02 0 -8 -16 -9 -13 -18 92
## 3 1997-01-03 -4 -6 -7 -6 -8 -9 93
## 4 1997-01-04 -3 -4 -5 -5 -6 -6 93
## 5 1997-01-05 -1 -3 -6 -4 -5 -7 100
## 6 1997-01-06 -2 -3 -4 -4 -5 -6 93
## 7 1997-01-07 0 -4 -9 -6 -9 -10 93
## 8 1997-01-08 0 -3 -7 -7 -7 -8 100
## 9 1997-01-09 0 -3 -6 -5 -6 -7 100
## 10 1997-01-10 -3 -4 -5 -4 -5 -6 100
## # … with 7,185 more rows, and 15 more variables: mean_hum ,
## # min_hum , max_hpa , mean_hpa , min_hpa ,
## # max_visib , mean_visib , min_visib , max_wind ,
## # mean_wind , max_gust , prep , cloud ,
## # events , winddir
Window (AKA analytic) functions
Joins, set operations, that’s pretty cool to have but that’s not all. Additionally, a large number of analytic functions are available in dplyr. We have the familiar-from-SQL ranking functions (e.g., dense_rank(), row_number(), ntile(), and cume_dist()):
# 5% hottest days
filter(daily_2016, cume_dist(desc(mean_temp)) % select(day, mean_temp)
## # A tibble: 5 × 2
## day mean_temp
##
## 1 2016-06-24 22
## 2 2016-06-25 22
## 3 2016-07-09 22
## 4 2016-07-11 24
## 5 2016-07-30 22
# 3 coldest days
filter(daily_2016, dense_rank(mean_temp) % select(day, mean_temp) %>% arrange(mean_temp)
## # A tibble: 4 × 2
## day mean_temp
##
## 1 2016-01-22 -10
## 2 2016-01-19 -8
## 3 2016-01-18 -7
## 4 2016-01-20 -7
We have lead() and lag():
# consecutive days where mean temperature changed by more than 5 degrees:
daily_2016 %>% mutate(yesterday_temp = lag(mean_temp)) %>% filter(abs(yesterday_temp - mean_temp) > 5) %>% select(day, mean_temp, yesterday_temp)
## # A tibble: 6 × 3
## day mean_temp yesterday_temp
##
## 1 2016-02-01 10 4
## 2 2016-02-21 11 3
## 3 2016-06-26 16 22
## 4 2016-07-12 18 24
## 5 2016-08-05 14 21
## 6 2016-08-13 19 13
We also have lots of aggregation functions that, if already provided in base R, come with enhancements in dplyr. Such as, choosing the column that dictates accumulation order. New in dplyr is e.g., cummean(), the cumulative mean:
daily_2016 %>% mutate(cum_mean_temp = cummean(mean_temp)) %>% select(day, mean_temp, cum_mean_temp)
## # A tibble: 260 × 3
## day mean_temp cum_mean_temp
##
## 1 2016-01-01 2 2.0000000
## 2 2016-01-02 -1 0.5000000
## 3 2016-01-03 -2 -0.3333333
## 4 2016-01-04 0 -0.2500000
## 5 2016-01-05 2 0.2000000
## 6 2016-01-06 2 0.5000000
## 7 2016-01-07 3 0.8571429
## 8 2016-01-08 4 1.2500000
## 9 2016-01-09 4 1.5555556
## 10 2016-01-10 3 1.7000000
## # … with 250 more rows
OK. Wrapping up so far, dplyr should make it easy to do data manipulation if you’re used to SQL. So why not just use SQL, what can we do in R that we couldn’t do before?
Visualization
Well, one thing R excels at is visualization. First and foremost, there is ggplot2, Hadley Wickham‘s famous plotting package, the realization of a “grammar of graphics”. ggplot2 predates the tidyverse, but became part of it once it came to life. We can use ggplot2 to plot the average monthly temperatures from Berkeley Earth for selected cities and time ranges, like this:
cities = c("Munich", "Bern", "Oslo")
df_cities % filter(city %in% cities, year(dt) > 1949, !is.na(avg_temp))
(p_1950 <- ggplot(df_cities, aes(dt, avg_temp, color = city)) + geom_point() + xlab("") + ylab("avg monthly temp") + theme_solarized())
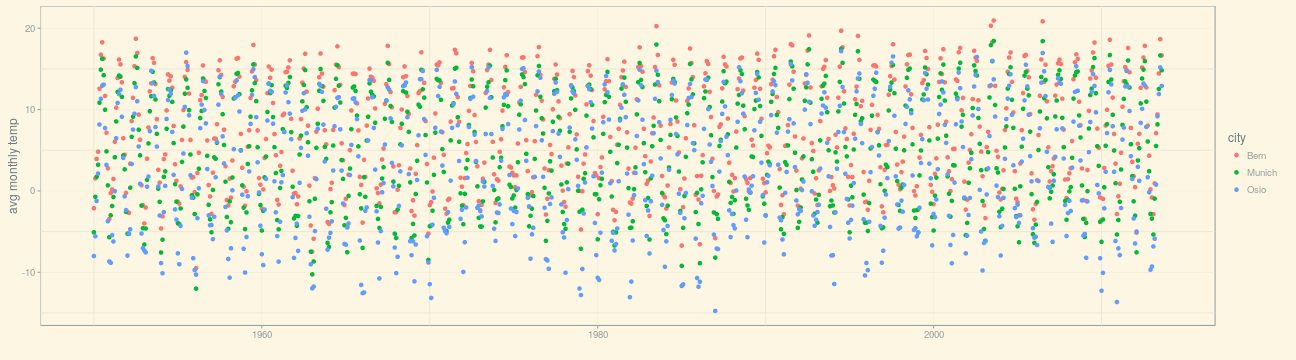
While this plot is two-dimensional (with axes time and temperature), a third “dimension” is added via the color aesthetic (aes (…, color = city)).
We can easily reuse the same plot, zooming in on a shorter time frame:
start_time <- as.Date("1992-01-01")
end_time <- as.Date("2013-08-01")
limits <- c(start_time,end_time)
(p_1992 <- p_1950 + (scale_x_date(limits=limits)))
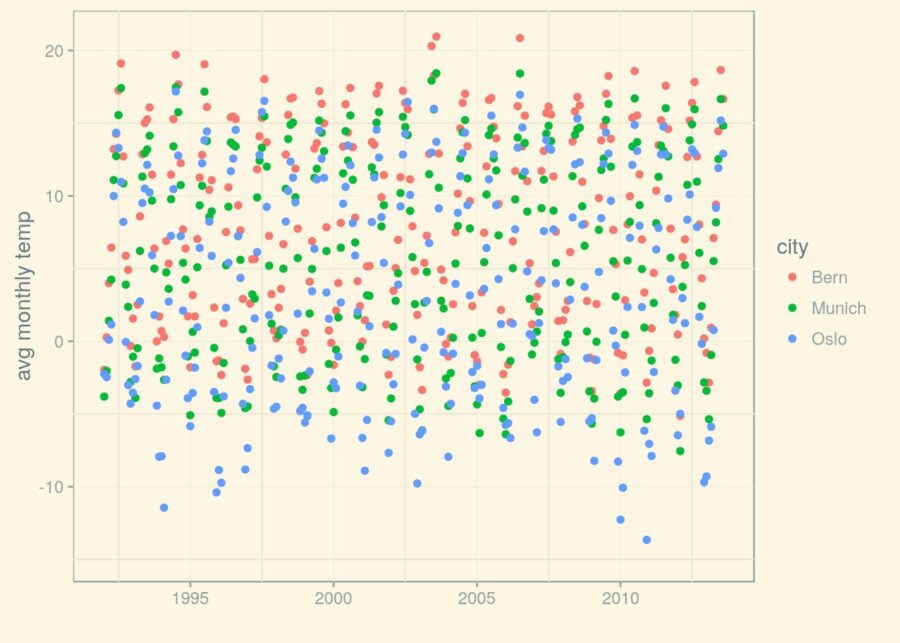
It seems like overall, Bern is warmest, Oslo is coldest, and Munich is in the middle somewhere.
We can add smoothing lines to see this more clearly (by default, confidence intervals would also be displayed, but I’m suppressing them here so as to show the three lines more clearly):
(p_1992 <- p_1992 + geom_smooth(se = FALSE))
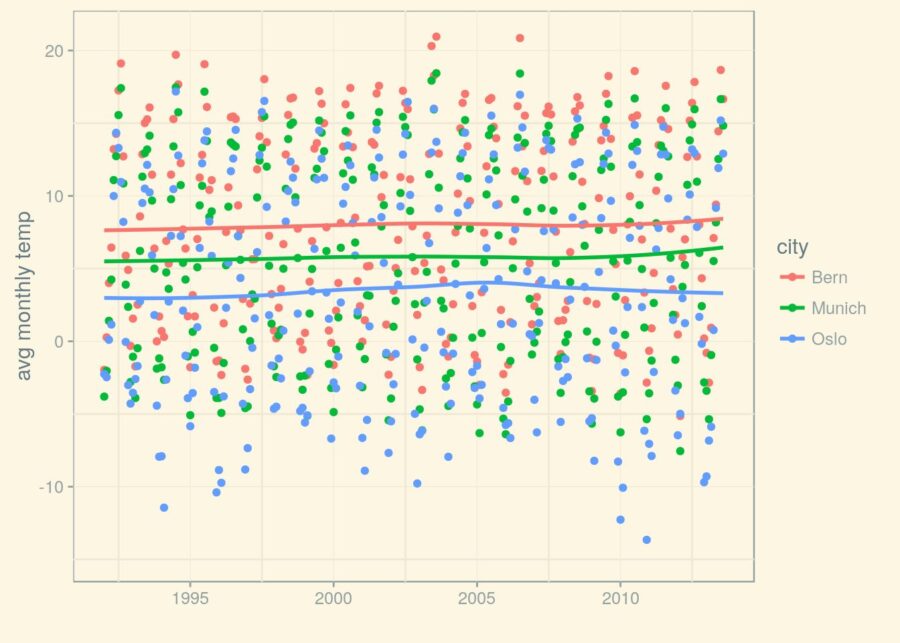
Good. Now that we have these lines, can we rely on them to obtain a trend for the temperature? Because that is, ultimately, what we want to find out about.
From here on, we’re zooming in on Munich. Let’s display that trend line for Munich again, this time with the 95% confidence interval added:
p_munich_1992 <- p_munich_1950 + (scale_x_date(limits=limits))
p_munich_1992 + stat_smooth()
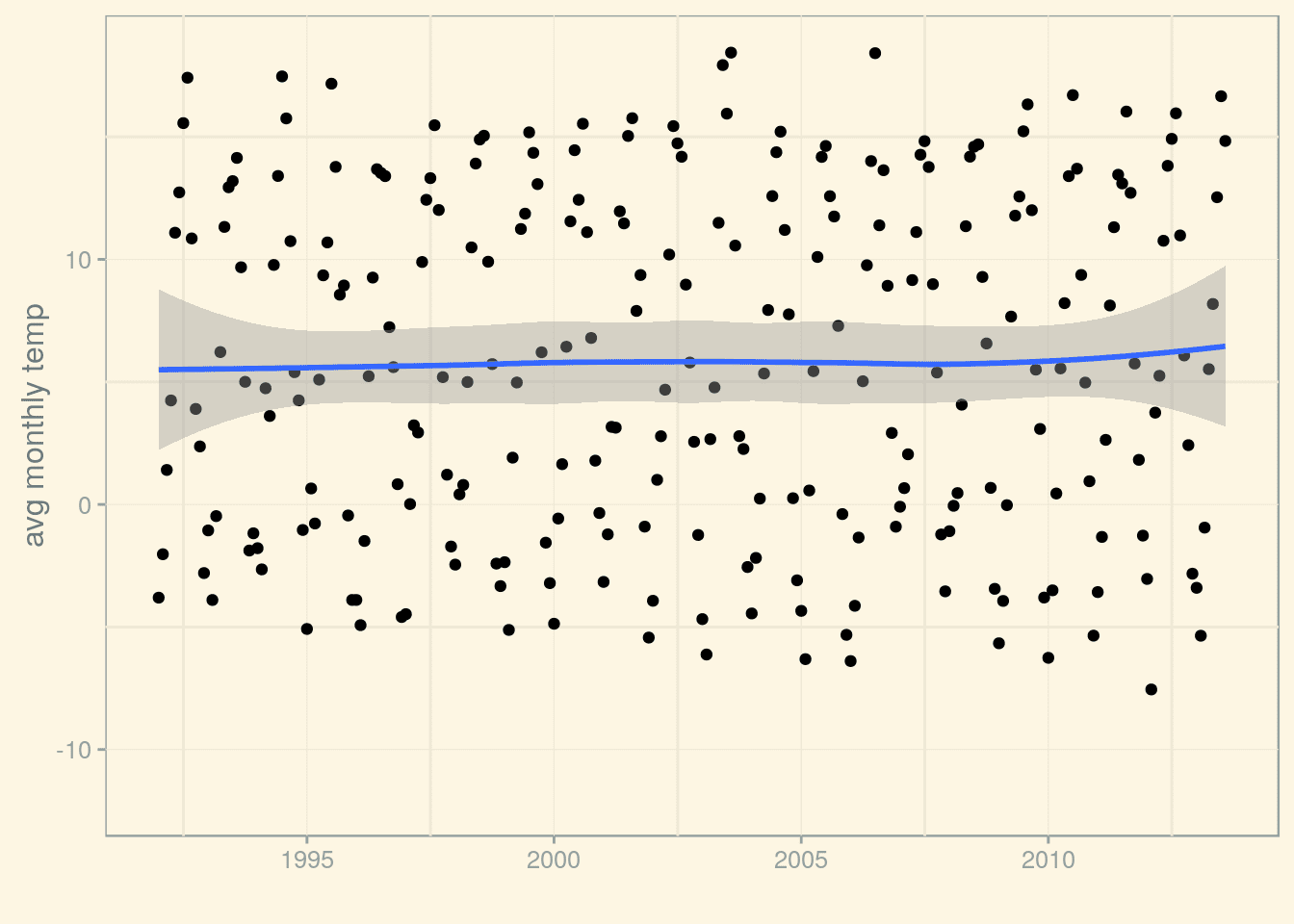
Calling stat_smooth() without specifying a smoothing method uses Local Polynomial Regression Fitting (LOESS). However, we could as well use another smoothing method, for example, we could fit a line using lm(). Let’s compare them both:
loess <- p_munich_1992 + stat_smooth(method = "loess", colour = "red") + labs(title = 'loess')
lm <- p_munich_1992 + stat_smooth(method = "lm", color = "green") + labs(title = 'lm')
grid.arrange(loess, lm, ncol=2) (p_1992 <- p_1950 + (scale_x_date(limits=limits)))
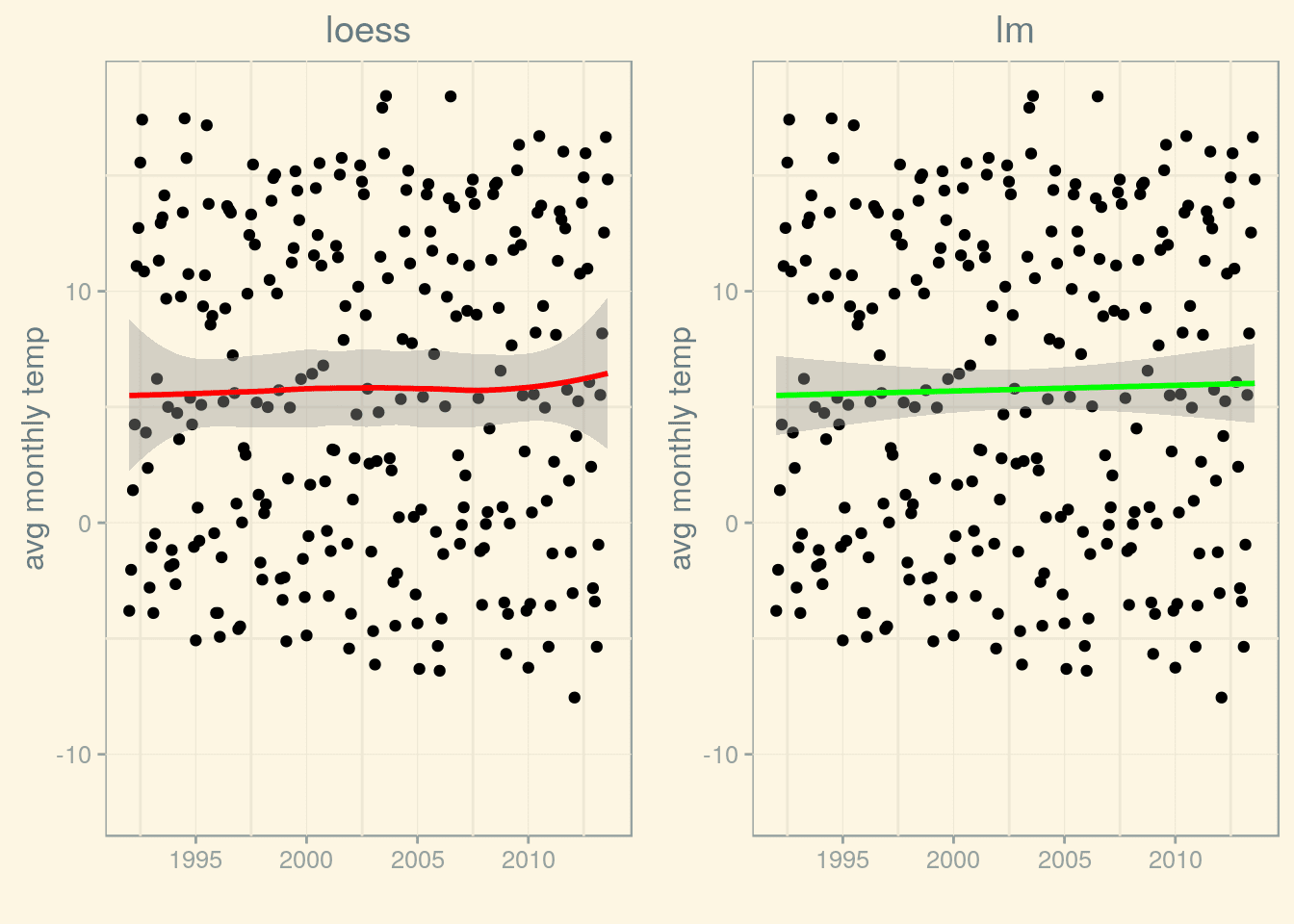
Both fits behave quite differently, especially as regards the shape of the confidence interval near the end (and beginning) of the time range. If we want to form an opinion regarding a possible trend, we will have to do more than just look at the graphs – time to do some time series analysis!
Given this post has become quite long already, we’ll continue in the next – so how about next winter? Stay tuned 🙂
Note: This post has originally been postedhere
