When people talk about Large Language Models, the most common topics are text summarization, text generation, and answering prompts with GPT. Yet, this is just the tip of the iceberg. What if the language has an unusual alphabet? In this article, I discuss creating meaningful, synthetic sentences — very long ones with millions of letters — in a peculiar language. Its alphabet has 4 letters: A, C, G, T. Each one represents a protein: adenine (A), cytosine (C), guanine (G), and thymine (T). This is the DNA language, and the long sentences are DNA sequences.
Patterns Found in DNA Sequences
Just like standard English, letter and word combinations are not random. Some happen frequently, and some not at all. Then, rare combinations may indicate a particular genetic condition, similar to typos in the English language. But the big difference is the absence of separators (commas, spaces, question marks), creating long, uninterrupted sequences of letters. In this context, a word is any combination of 2, 3 or more consecutive letters. Because of this, all words overlap.
In some sense, DNA synthetization is somewhat easier than generating English text. Thus, this could be a first project for professionals starting with LLMs. Yet, there are long-range autocorrelations and non-probabilistic rules. For instance, how and where do I insert the correct word that identifies the gender of a human being? I will not answer these questions here. Instead, I focus on rather short-range patterns. In the end, it is not that much different from standard LLMs focusing mostly on adjacent tokens and autoregressive predictors. In both cases, word embeddings are a critical component.
Figure 1 shows three DNA subsequences: a real one, a synthetic one, and a random sequences of letters A, C, G, T. Can you identify patterns in the two top ones?

The Training Set
The training set consists of a number of human DNA sequences, and it is publicly available. The original project consisted in classifying different subsequences showcasing different genetic traits, based on the patterns and words found, including their statistical distributions. I grouped the data, producing a sequence with a few millions letters. In addition to the 4 letters A, C, G, T, there were a number of occurrences of the letter N, presumably representing missing data.
The training set and Python code are on my GitHub repository. You can access them by downloading the technical document available as paper #34, here. An interesting ethical question is about privacy: the individual subsequences may be enough to identify the corresponding persons, and thus the diseases that they might have. Thankfully, the synthetic DNA should make this reverse-engineering — matching DNA against existing databases — impossible.
Note that the technology discussed here also works in other contexts. For instance, to synthesize drugs or molecules.
Synthesizing DNA Sequences
The algorithm has two steps. First looking at pairs of adjacent words and compute occurrences and conditional probabilities. Then sequentially generate new words based on the last generated word and on the probabilities in question. In the end, you end up with a large table of word embedding, each embedding having hundreds or thousands of components (related words), just like in standard LLM.
More specifically, it works as follows. Look at a DNA word S1 consisting of n1 consecutive letters, to identify potential candidates for the next word S2 consisting of n2 symbols. Then, assign a probability to each word S2 conditionally on S1, and use these transition probabilities to sample S2 given S1, then move to the right by n2 letters, do it again, and so on. Eventually you build a synthetic sequence of arbitrary length. There is some analogy to Markov chains and autoregressive processes.
An interesting application consists of generating synthetic sequences — thus artificial human beings — then produce pictures of the artificial people in question based on their artificial DNA. To do this, you may infer physical appearances based on the DNA: eye and hair color, nose shape, and so on.
Evaluating the Quality of Synthetic DNA
To evaluate the results, I compared the frequencies of a large number of words, both in a synthetic and real sequence. I used the Hellinger distance, returning a value between 0 (best) and 1 (worst). Figure 2 is a QQ plot, with each dot representing a word. The X-axis represents the frequency of the word in question in the synthetic data, while the Y-axis represents its frequency in real DNA.
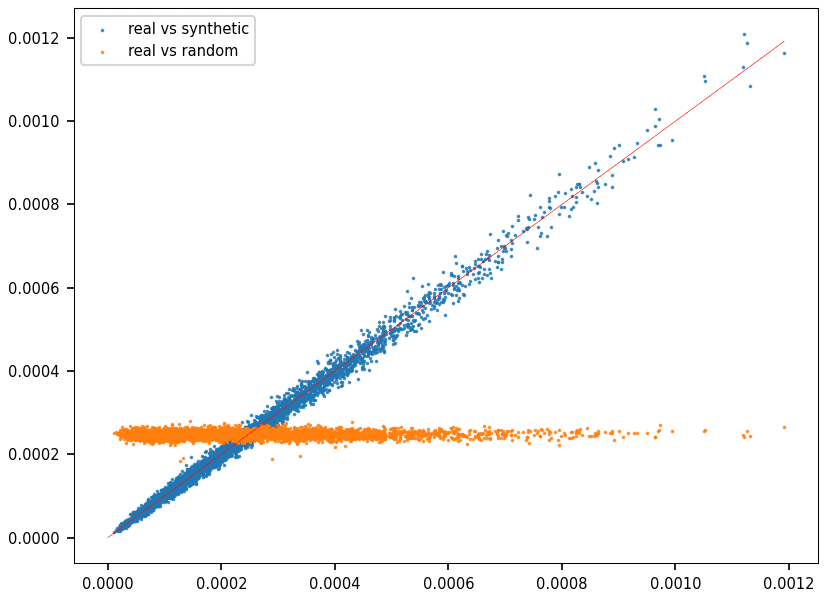
Clearly, if you look at Figure 2, DNA sequences are anything but random. The synthetic DNA replicates the correct distribution, while the random DNA is completely off. However, the evaluation metric depends on the length of the words used. In this case, all 4000 selected words had 6 letters. A QQ plot with ten thousand 8-letter words is featured in paper #34, here. The fit is not as great a in Figure 2. In general, these QQ plots do not capture long-range interactions.
Author

Vincent Granville is a pioneering GenAI scientist and machine learning expert, co-founder of Data Science Central (acquired by a publicly traded company in 2020), Chief AI Scientist at MLTechniques.com and GenAItechLab.com, former VC-funded executive, author and patent owner — one related to LLM. Vincent’s past corporate experience includes Visa, Wells Fargo, eBay, NBC, Microsoft, and CNET.
Vincent is also a former post-doc at Cambridge University, and the National Institute of Statistical Sciences (NISS). He published in Journal of Number Theory, Journal of the Royal Statistical Society (Series B), and IEEE Transactions on Pattern Analysis and Machine Intelligence. He is the author of multiple books, including “Synthetic Data and Generative AI” (Elsevier, 2024). Vincent lives in Washington state, and enjoys doing research on stochastic processes, dynamical systems, experimental math and probabilistic number theory. He recently launched a GenAI certification program, offering state-of-the-art, enterprise grade projects to participants.
