
Year 1994 – I stood helpless while my dear friend’s Father was going through traditional chemotherapy which is known to affect all cells indiscriminately. That was the best the doctors were able to do at that time notwithstanding the fact that each cancer is different caused by an uncontrollable mutation of bad cells and the ideal combination or sequence of drugs should stop these bad cells from reproducing whilst preserving the good ones. Those were just my initial year into the IT industry and made me feel helpless as an engineer for not being able to contribute any thoughts towards the brave efforts from Oncologists & Radiologists.
Two decades later, whilst I was shifting gears from corporate into the startup world of AI, I got intrigued by an article from Dr Pedros Domingos (University of Washington) on the potential power of machine learning algorithms in the field of medicine. These machine learning algos were probably the same as discovered more than half a century back but their full potential and predictive powers were being unleased now with the availability of unlimited compute capacity via cloud at a fraction of cost. With that started my quest for finding a “bespoke cure” (as each instance of cancer is difference) for cancer by cross leveraging the powers in the field of artificial intelligence, genome sequencing, cloud computing, molecular biology & drug trials.
The Human Cell
To find the root cause of any cancer we have to zoom inside the anatomy of a human body cell which is a highly complex non-linear structure the functioning of which is dictated by a unique set of genetic instructions (the “Genome”). These genetic instructions per cell are made up of 3.2 billion base pairs of negatively charged DNA twisted around and bonded by a set of positively charged histone proteins. The sequence of these chemical/nucleotide base pairs – also known as the “genetic letters”: The As (Adenine), the Cs (Cytosine), the Gs (Guanine) and the Ts (Thymine) determines the unique chemical code of the DNA and the cell. And an average Human body is made up of 50 trillion of these cells each with its unique Genome or sequence of the 3.2 billion nucleotide base pairs / genetic letters. So it’s just impossible for the human mind to comprehend this complex dataset “if printed out the 3.2b letters in your genome would take a century to recite if we recited at one letter per second for 24 hours a day” [https://www.yourgenome.org/facts/what-is-a-genome ] .
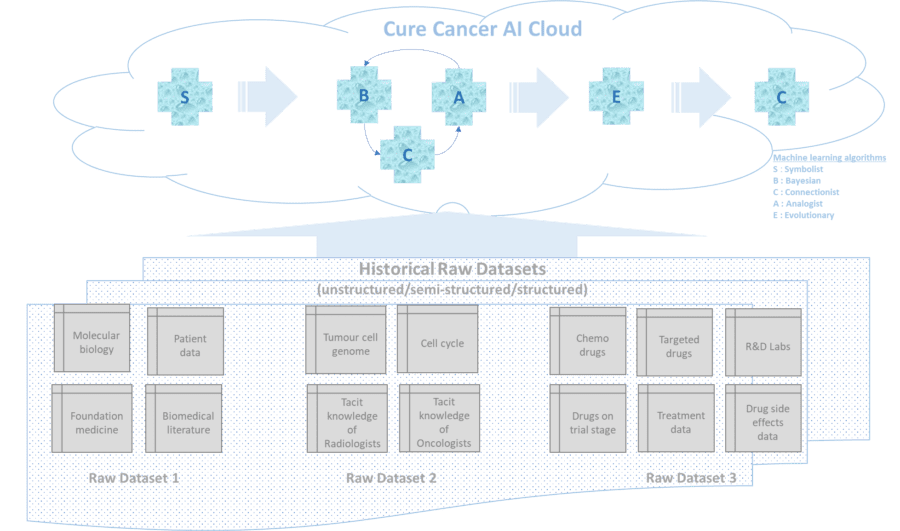
However, this complex unstructured dataset of molecular biology / biomedical literature can easily be digested by an AI engine (powered by unparalleled compute capacity from the cloud). Lets term this as “Raw Dataset 1” for our Cure Cancer AI Cloud.
The Genes & their interactions
Within the DNA of each cell are 30,000 genes which control the growth, division and life span (the cell cycle) of the cell. The genes make proteins which act as messengers for the cell and bind the nucleotide base pairs of DNA in the cell. If for some reason one or more genes in a cell mutates i.e. changes – it starts making abnormal protein/s which in turn render distorted information through the cell’s metabolic pathways causing the cell to divide uncontrollably and become cancerous / bad cell. Some of these mutations are suppressed / regulated by DNA repair genes or tumour suppressor genes within the cell so the cell gets into a self-healing regime. However, some instances of multiple gene mutations may lead to concentration of mutated bad cells or a cancerous tumour. Each cancer is different due to the varying sequence of the 3.2b chemical base pairs (As,Cs,Gs,Ts) in the DNA of the cell. Genome sequencing can help in determining the exact sequence of these base pairs based on which starts the key process of identifying the ideal sequence or combination of drugs to stop the reproduction of those cancerous cells. With the ever-increasing compute capacity from the cloud – the cost of genome sequencing has drastically reduced from $300m to less than $1000.
Lets consider the cell cycle – the genes – possible mutation/s – inter gene interaction/regulation – genome sequencing and associated patient data as “Raw Dataset 2” for our Cure Cancer AI Cloud.
The Chemo & Targeted Drugs and the Drug ecosystem
There are 100s of standard chemo drugs and targeted drugs which are used to cure cancer and its vital to have a thorough understanding of their chemical structures and interaction with the metabolic structure of the human cell to aid in maximising their effect on cancerous cells and minimising their side effects on healthy cells. The chemo drugs may attack different parts of the cell like the proteins (Topoisomerase inhibitors, Anthracyclines, etc) , the DNA (antimetabolites, Alkylating drugs, etc), the genes (Mitotic inhibitors, etc). However, they can have serious side effect due to their action on healthy cells for example Alkylating agents effect the new blood cells in the bone marrow, a high dose of Anthracyclines may inflict a permanent damage to the heart, etc.
Along with drugs available to hospitals there are drugs being tested or trialled across thousands of R&D Labs across the globe and the 200+ global cancer research groups .
Lets consider all the chemo and targeted drugs in prescription and trial stages – their chemical function on mutated cells – their side effects as “Raw Dataset 3” for our Cure Cancer AI Cloud.
The Cure Cancer AI Cloud
The world of AI is studded with machine learning algorithms each with their own strengths and limitations. Hence it is critical to apply the right combination and sequence of these algos to the raw datasets highlighted above. The first step would be to simulate a real-time multi-dimensional neural model of the cell, the cell cycle, the genes & their possible mutations and inter gene regulation in cancerous cells by combining disparate set of knowledge banks from biomedical literature (raw dataset1) and genome sequencing (raw dataset 2) and tacit knowledge of Oncologists/Radiologists/etc. The Symbolist class of machine learning algos (via decision trees, inverse deduction) are best suited for this basic first step as they can just learn on the fly by combining these staggered pieces of knowledge/datasets to simulate the draft real-time multi-dimensional view of the required neural graph with millions/billions of nodes wherein each node represents either a DNA or gene or protein or a nucleotide base pair/genetic letter – The As , the Cs, the Gs, the Ts, etc.

The resultant neural graph can give a clear picture to the Oncologists, with 60% to 75+% prediction levels, on the location and evolving mutation of the cancer cells and associated cell cycle. However, the probability % is lower due to limitation of Symbolists on incomplete or contradictory datasets (raw datasets 1 & 2 with reference to this paper) and the insights generated by them are mostly qualitative.
At this stage we need to trigger the combo of Bayesian and Connectionist (also known as deep learning) class of machine learning algos. The Bayesians (via probabilistic inference) thrive on partial/incomplete/noisy/contradictory datasets and can firm up the draft neural graph generated by the Symbolist as above to substantially higher accuracy & probability levels. And in parallel, the Connectionists (via back propagation) can generate quantitative view / strength of the billions of connections (inter-gene interaction, etc) in the neural graph to give a deeper insight into how the mutations are evolving. The combo action of Bayesian and Connectionist algos on the output of Symbolists will give us a firm grip/insight on the changing metabolic structure of the cells to 90+% confidence levels. This can further be cross-corelated to patient datasets with similar or superficially different symptoms by Analogists class of machine learning algos which (via support vector machine) can do self-learning by as little as one example and draw analogy/essential similarities between very different symptoms.
Now will be the time to fire up the Evolutionary class of machine learning algo which (via genetic search) can cross-corelate the effect of drugs (from raw dataset 3) on the evolving metabolic structure of the cells to get a deeper insight as to which combination of drugs (across prescription and trial stages in the global drug eco-system) and what sequence of the same can stall the mutation of the cancerous/tumour cells with minimal or no side effect on the healthy cells. And via their crossover function the Evolutionary algos can infer a set of hypothesis on designer drugs and their sequence for bespoke cure based on the insights into the genome sequence of the tumour cells. However, we need to check the strength of each set of hypothesis and this is where we spawn off the Connectionist set of algos again to help us proceed deterministically with 90%+ confidence level on the bespoke cure for a specific instance of cancer.
The proposed “Cure Cancer AI Cloud” in this open paper may not be 100% quintessential but is a sincere effort to invite an open collaboration from the AI fraternity, the chemo & targeted drug ecosystem, R&D trial labs, the Oncologists, Radiologists and cancer research organisations towards the noble cause of curing cancer and save precious life.

About the Author:
Mukul Gupta is an entrepreneur with 25 years’ experience spanning across AI, Telecom, Oil & Gas, Utilities, Education, Insurance, FS & Retail sectors handling various Executive, Regional sales, Global Account Leadership & SI roles across Europe, Australia, NZ, US & India. Awarded a US Patent for ‘Collaborative Customer Service Model’
Mukul has been an active speaker & panellist in International Forums at Europe, US, Australia & China. His thought leadership in the industry has been acknowledged by the Millennial media (The Market Mogul), leading publications (Power Engineering International, Capacity, Mycustomer.com, Lightreading, Total Telecom, Mobile News), analysts (Gartner, Ovum) and international organizations & forums (IEC, IPDR, Marcus Evans, ACIF, Quest, NOA UK, BITE).
