Objective
In this tutorial, we shall take a journey together to explore the structure of the DrugBank database. We will observe how the drugs information is structured within DrugBank’s XML database and see how this information can be retrieved using R. Our main purpose here is parsing the database from its containing XML file. Let us begin!
What is DrugBank?
DrugBank is a comprehensive, freely accessible, online database containing information on drugs and their targets. As both a bioinformatics and a cheminformatics resource, DrugBank combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information.
As both a bioinformatics and a cheminformatics resource, DrugBank combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information.
The DrugBank XML file
Below is what the XML file looks like on the inside. As we can see, there is a single <drugbank> node and, within it, lie thousands of <drug>nodes (which we talk about in greater detail in the following section). These nodes contain the information of the many drugs that constitute the DrugBank database.

To follow along with the code in this tutorial, you may download and use this XML file. It is a dummy XML database file that contains only a single drug record (i.e. a single <drug> node). Alternatively, you may instead download and use the entire DrugBank XML database file which is available here.
The <drug> node (and its children)
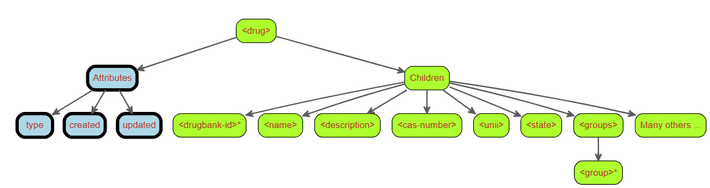
Each drug in the database is represented by a drug node which contains, along with its associated attributes, many children nodes with varying names, properties and (more importantly) structures.
Below is a portion of a <drug> node as an example.

What we see in the example above is the following:
- The
strong>drug>node has three attribute values:type: the drug type (i.e.biotechorsmall molecule)created: the date when this particular drug was createdupdated: the date when this particular drug was last updated
- The
strong>drug>node has many children:- One of the children nodes,
strong>drugbank-id>, can appear more than once under the<drug>node. - Some of the children nodes consist of a single value (
strong>name>,strong>description>,strong>cas-number>,strong>unii>,strong>state>). - Other children nodes have multiple children nodes themselves (
strong>groups>).
- One of the children nodes,
- The
<drug>node above includes many other children that, for the moment, have been left out for the sake of simplicity. As we will see, some of those other children are much more complex than the ones shown above. We will find that these children vary greatly in structure; some of them simply contain a single value while others may contain multiple children nodes or sometimes even deeper hierarchies of children nodes within them. All these children nodes may have their own attributes as well (not just the drug node).
Right now, our current conceptual understanding of the structure of a strong>drug> node looks something like the figure below.