Introduction
As we see COVID19 affected 1.5M people globally and 100k deaths, the worldwide AI research community are trying help medical community whatever the way we could. Many of medical questions are leads to research papers for answers. These questions are suitable for text mining, and developing text mining tools to provide insights on these questions.
In this blog, I want to share how you can use BERT based embeddings pre-trained model can be used to build a text-based Question and Answering tool for fining answers related to Coronavirus from COVID19 research papers repository.
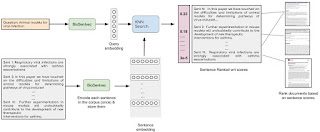
When I was with IBM, I involved a similar solution to IBM Risk Insights, which search for supply chain risks from the news articles, tweets, and weather data. Based on the base idea from the Risk Insights project and Biology knowledge domain for embeddings, I developed this model. With this model, users can search for research papers to find out and outputs related articles and which paper, under which section the user could find answers for the input question. Also, it displays the related paragraphs as output. It returns top article with highlighted answer sentences from the articles. My code is also uploaded to my Github
Data
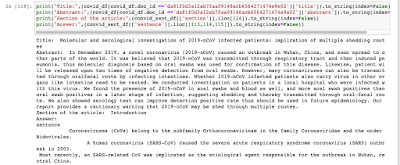
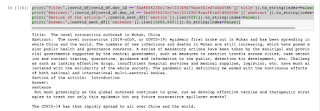
In response to the COVID-19 pandemic, the White House and a coalition of leading research groups have prepared the COVID-19 Open Research Dataset (CORD-19). CORD-19 is a resource of over 52,000 scholarly articles, including over 41,000 with full text, about COVID-19, SARS-CoV-2, and related coronaviruses. This dataset is publicly available in Kaggle’s COVID-19 Open Research Dataset Challenge (CORD-19). I used only articles in the JSON format for this model. The dataset has 7865 articles in JSON format which contains 64,000 unique sections and 1.1M sentences. Also, it has articles metadata such as data published, authors, title, and abstract.
Sentence Embedding
This solution primary for the medical domain so I used BioSentVec pre-trained model for embedding. BioSentVec created biomedical word and sentence embeddings using PubMed and the clinical notes from MIMIC-III Clinical Database. Both PubMed and MIMIC-III texts were split and tokenized using NLTK. We also lowercased all the words. More details and a pre-trained model can use accessed here.
Architecture
This solution is a type of Question Answering model. It is a retrieval-based QA model using embeddings. The basic idea of this solution is comparing the question string with the sentence corpus, and results in the top score sentences as an answer. I create a vector representation of each sentence using a pre-trained BioSentVec embedding model and KNN to find the answer sentences.
Preprocess
Below are the steps I took to preprocess the data. Again, you can access full code in my GitHub.
● Read JSON files from dataset
● Parse JSON files
● Remove hyperlinks, references
● Extract section details and split article by sentences using NLTK’s sent_tokenize method.
● Generate pandas data frame for model
Embedding sentences
Loading the BioSentVec pretrained model
model_path = ‘BioSentVec_PubMed_MIMICIII-bigram_d700.bin’
model = sent2vec.Sent2vecModel()
try:
model.load_model(model_path)
except Exception as e:
print(e)
print(‘model successfully loaded’)
Vectorize the sentence corpus with BioSentVec model
embs = model.embed_sentences(convid_sent_df[‘sentence’])
KNN and Ranking
The k-Nearest Neighbors algorithm (KNN)t is a very simple technique. First, I loaded entire vectorized sentences into the model as training. When I need to find the answer, we need to send a vectorized question string as input to the model and the knn model outputs the most similar records from the training sentence corpus with the score. From these neighbors, a summarized answer is made. A similarity between records can be measured in many different ways. I used default here.
from sklearn.neighbors import NearestNeighbors
nbrs = NearestNeighbors(n_neighbors=2, algorithm=’ball_tree’).fit(embs)
Test the Model
Using trained kneighbors model passing the question. As “ What is the physical science of the coronavirus’
emb = model.embed_sentence(‘Physical science of the coronavirus’)
distances, indices = nbrs.kneighbors(emb)
Output
Conclusion
Using BioSentVec embedding and KNN algorithms is not the only solution, here are few alternative approaches.
● We can develop a custom model based on predefined keyword (fuzzy keywords) domain dictionary and redefined rules using regular expression for building a rule-based semantic search tool. This kind of model works very well with a narrow knowledge set and can be implemented quickly.
● We can develop a quick Question Answering model using BERT based pre-trained Sentence Transformer models.
● And many more options
I am planning to try a similar question answering model with twitter streaming data.
Final Note: I am not a heather professional to comment on the output provided by the model and this article purpose to illustrate the possible solution.
References:
● This solution primarily based on the BioSentVec pre-trained morel big thanks for the developers and the original BERT model developed.
● Dataset downloaded from Kaggle.com. https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge