This article was written by John Mount.
Introduction
In this note we will show how to speed up work in R by partitioning data and process-level parallelization. We will show the technique with three different R packages: rqdatatable, data.table, and dplyr. The methods shown will also work with base-R and other packages.
For each of the above packages we speed up work by using wrapr::execute_parallel which in turn uses wrapr::partition_tablesto partition un-related data.frame rows and then distributes them to different processors to be executed. rqdatatable::ex_data_table_parallel conveniently bundles all of these steps together when working with rquery pipelines.
The partitioning is specified by the user preparing a grouping column that tells the system which sets of rows must be kept together in a correct calculation. We are going to try to demonstrate everything with simple code examples, and minimal discussion.
Keep in mind: unless the pipeline steps have non-trivial cost, the overhead of partitioning and distributing the work may overwhelm any parallel speedup. Also data.table itself already seems to exploit some thread-level parallelism (notice user time is greater than elapsed time). That being said, in this note we will demonstrate a synthetic example where computation is expensive due to a blow-up in an intermediate join step.
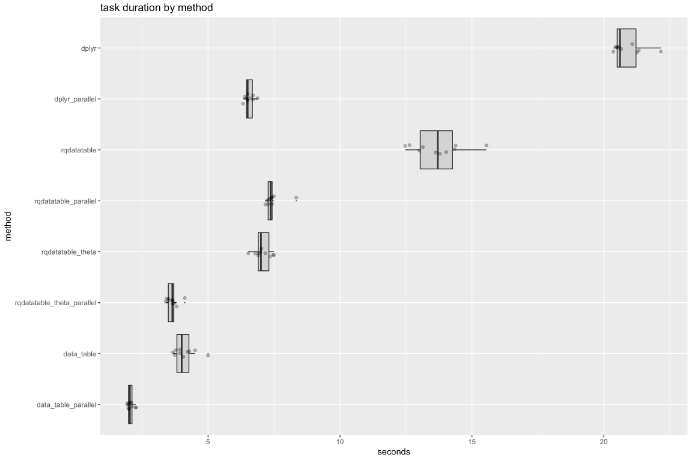
Conclusions
The benchmark timings show parallelized data.table is the fastest, followed by parallelized dplyr, and parallelized rqdatatable. In the non-paraellized case data.table is the fastest, followed by rqdatatable, and then dplyr.
A reason dplyr sees greater speedup relative to its own non-parallel implementation (yet does not beat data.table) is that data.table starts already multi-threaded, so data.table is exploiting some parallelism even before we added the process level parallelism (and hence sees less of a speed up, though it is fastest).
rquery pipelines exhibit superior performance on big data systems (Spark, PostgreSQL, Amazon Redshift, and hopefully soon Google bigquery), and rqdatatable supplies a very good in-memory implementation of the rquery system based on data.table. rquery also speeds up solution development by supplying higher order operators and early debugging features.
In this note we have demonstrated simple procedures to reliably parallelize any of rqdatatable, data.table, or dplyr.
To read the rest of the article, with an example and source code, click here. This material is also available on GitHub, here.
