Introduction
How often are you in a situation where you have 2 alternatives either yes or a no, black or a white, and so on. These are instances where you ‘classify’ your scenario into only two solutions, a number of solutions may vary but usually, they are two solutions. This is what we call as ‘Classification’ we classify the outcomes in a set number of instances usually two. This week at The Datum we have how can we use Neural Networks as the classification model. And once we have the model in hands we will go about prediction using the model and lastly, we will evaluate our model and predictions for its rightness.
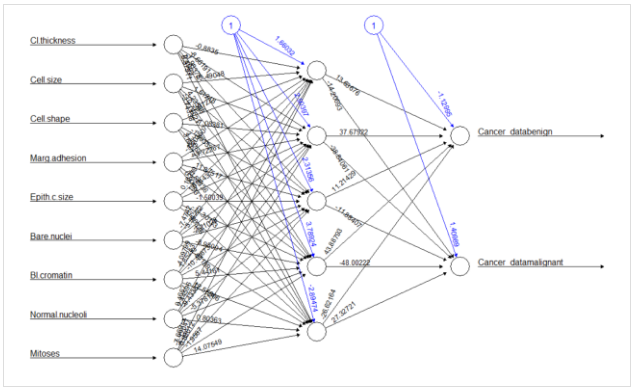
Model Brief
Today we have a very interesting model to build which can classify breast cancer. Cancer is uncontrolled multiplication of cells. And breast cancer is caused due to the multiplication of cells in the mammary gland that are transformed into malignant cells. They have the ability to detach themselves from the tissues which they are formed and invade into the surroundings due to multiplication. Breast is formed from multiple types of cells that form the breast but, the most common breast cancers are from glandular cells or from those forming the walls of the ducts.
The objective of this model today is to classify the number of benign and malignant classes which form the two most common type of breast cancer. For this model, we will be using the data already present in R named BreastCancer which is available in mlbench package. Using this data we will classify benign and malignant types of breast cancer using neural networks as a classifier.
Model Data
The data as mentioned is readily available in R library of mlbench and each variable except the first, is in the form of 11 numerical attributes with values ranging from 0 through 10, with some missing values as well. Following are the data fields of this data set.
$Id: Sample code number
$Cl.thickness: Clump thickness
$Cell.size: Uniformity of cell size
$Cell.shape: Uniformity of cell shape
$Marg.adhesion: Marginal adhesion
$Epith.c.size: Single epithelial cell size
$Bare.nuclei: Bare nuclei
$Bl.cromatin: Bland chromatin
$Normal.nucleoli: Normal nucleoli
$Mitoses: Mitoses
$Class: Class
With good amount of information on the model brief and data we are good to implement the Neural Network Classifier model in R. To have an in-depth understanding of Neural Network concepts and functioning visiting the blog Prediction Analysis with Neural Networks and Linear Regression is recommended (here), which has all the information needed to understand the working of Neural Networks. To have complete access to this blog with entire listings, data and building the algorithm visit, read here.
