In this short blog, we’ll review the process of taking a POC data science pipeline (ML/Deep learning/NLP) that was conducted on Google Colab, and transforming it into a pipeline that can run parallel at scale and works with Git so the team can collaborate on.
Background
Google Colab is pretty straightforward, you can open a notebook in a managed Jupyter environment, train with free GPUs, and share the drive notebooks, it also has a Git interface to some extent (mirroring the notebook into a repository). I wanted to scale my notebook from the exploration data analysis (EDA) stage to a working pipeline that can run in production. It was also important to have better Git integration for storing the artifacts and collaborating with the team. Ploomber allowed me to achieve it, while also accelerating the development stage by building incrementally tasks that changed.
How does it work?
I started by bootstrapping my Colab with a few dependencies (pip install ploomber). I was using the first-pipeline example provided by Ploomber – Getting data, cleaning it, and visualizing it. The pipeline structure was specified in the pipeline.yaml file which is composed of tasks that can be functions, .py scripts or notebooks (nodes in my graph), and its products, each product can be many things like raw/clean data, executed notebooks, HTML reports, etc.
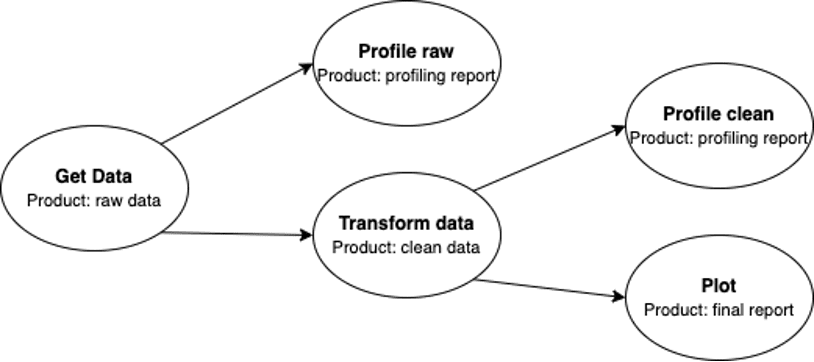
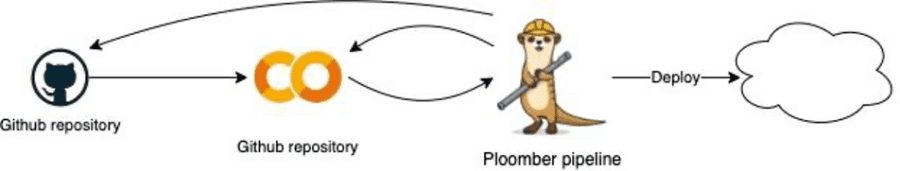
In the diagram below you can see that Ploomber and my Colab are in sync, loaded from and to the Git repository, and I can change the files that compose the pipeline directly from Colab. In case I’m done writing my POC I can deploy to the cloud with a single command, for instance to Airflow, Kubernetes, or AWS.
Benefits
This addition allowed me to develop rapidly and move from a monolith notebook to a scalable pipeline. I later pushed all of the code to GitHub so now we can collaborate as a team on the work and work simultaneously – only the real code differences are being saved. In addition, my notebook isn’t running sequentially, which means faster runs via parallelization. This also standardizes the work so if we ever want to replicate this project to a different business use case (which is kind of having a template) it’ll require less maintenance, and the learning curve will be significantly shorter.
What is Ploomber?
Ploomer (https://github.com/ploomber/ploomber) is an open-source framework to help data scientists develop data pipelines fast and deploy them anywhere. By using it directly from notebooks it allows you to skip weeks-long refactoring and focus on the modeling work and not infrastructures. We have a really active community which you can check out here.
Thanks for reading!

